Neural networks for taxon prediction
These two trees are topologically identical. One of them is a river system creating some of the most fertile land on earth, supporting a population of over 300 million people. And the other is a phylogenetic tree of the An. gambiae complex. One of them is also tattooed on my arm. extra points if you can guess the rivers.
Anopheles gambiae; a complex world
🔗The major malaria mosquito Anopheles gambiae, is a species complex of at least seven recognised species, and at least four additional cryptic taxa. These taxa have varying contributions to malaria transmission, with An gambiae s.s and An. coluzzii considered the most important vectors, due to their anthropophilic nature, or in other words, their preference for human blood. It is estimated that the ancestor of An. gambiae and coluzzii diverged from the rest of the complex approximately 2 million years ago.
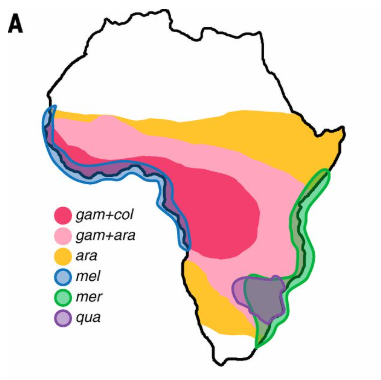
An. arabiensis on the other hand, shows more flexible host preferences but remains an important vector, particularly in drier areas, where it tends to dominate. Meanwhile, An. merus and An. melas are salt-tolerant coastal species and minor vectors. An. quadriannulatus is zoophilic (it prefers to feed on other animals) and is thought to play virtually no role in malaria transmission despite being competent to carry the parasite.
Despite these important phenotypic differences, these species are morphologically identical - you can't tell them apart even under a microscope. Traditionally, researchers have relied on molecular methods to identify members of the An. gambiae complex. The gold standard has been PCR-based assays targeting the ribosomal DNA intergenic spacer regions (IGS), developed by Scott et al. in 1993 and later with the SINE200 method (Santolamazza et al., 2008). Importantly, both methods rely on single genetic loci, which can miss hybridization between species, and these methods cannot detect cryptic taxa, and in those cases provide incorrect species assignments.
Neural networks for taxon prediction
🔗I've long been intrigued by the potential of machine learning to solve biological problems. For our recent development of a platform for targeted genomic surveillance of vectors, we needed to be able to identify species within the gambiae complex. The amplicon panel itself, originally designed long before my entry into the field by collaborators at the Sanger institute, was built for discriminating between An. gambiae and An. coluzzii, but not necessarily other species. I had been trying out some traditional machine learning methods, but with Large language models (LLMs) demonstrating remarkable capabilities in so many fields, I thought it was finally time to code up a neural network of my own.
Neural networks are computing systems inspired by the human brain's architecture. They consist of interconnected nodes ("neurons") organized in layers - an input layer that receives data, one or more hidden layers that process the information, and an output layer that produces the final result. Each connection between neurons has an associated weight, determining how much influence one neuron has on another. The network "learns" by adjusting these weights through a process called backpropagation, guided by a loss function that measures how far predictions deviate from known values.
Using free GPUs provided in Google Colab, alongside the TensorFlow and Keras libraries, I trained the network on ancestry informative markers within whole-genome sequence data from the Anopheles gambiae 1000 Genomes Project (Ag1000G). The neural network performed exceptionally well, achieving 100% accuracy in classifying all five species in the complex with sufficient numbers of samples: An. gambiae, An. coluzzii, An. arabiensis, An. melas, and the recently described Bissau molecular form. You can find the notebook for deep learning taxon prediction here.
Four SNPs to Rule Them All, and in the Darkness, Predict Taxa
🔗So, pretty amazing that a neural network can classify 5 species with 100% accuracy. Or is it? well, it turns out that neural nets are kind of overkill for this problem. In fact, what I found in the targeted surveillance paper, was that a simple decision tree using only four ancestry informative markers (AIMs) could predict the same taxa to a very high level.
The decision tree (Figure 3) showed remarkable accuracy for most species: An. gambiae (F1 = 0.995), An. coluzzii (F1 = 0.995), An. arabiensis (F1 = 1.000), and An. melas (F1 = 1.000). Although these SNPs were selected to distinguish between gambiae and coluzzi, it seems that they get fixed in different ways in the various species, allowing us to use these combinations to predict taxa. It performed slightly less well for the Bissau form (F1 = 0.849), likely because this cryptic taxon harbors less consistent AIM genotypes compared to the other species.
Figure 3. A simplified representation of a decision tree trained on four AIMs to predict species in the Anopheles gambiae complex
And back to my opening question - the right image depicts the Five Rivers of Punjab; the Jhelum, Chenab, Ravi, Beas, and Sutlej, tributaries of the Indus (admittedly, not in their correct orientation). The word Punjab literally means 'land of the five (panj) waters (ab)' The similar topology between these two systems - one representing my scientific work, and the other an ancestral home - was an intriguing reminder of the unexpected connections we can find in nature.
More Posts
Browse all posts